FAD-ETC-RBF Method
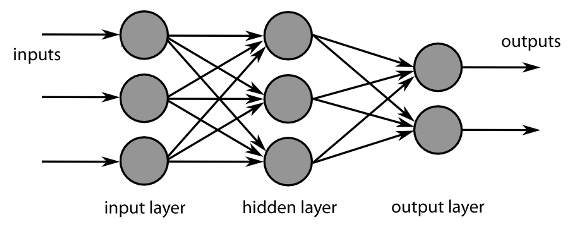
This study focused on identifying FAD binding sites in electron transport proteins. This figure illustrates a flowchart of the study, which included three subprocesses in each phase: data collection, feature set generation, and model evaluation. According to this flowchart, we developed a novel approach that is based on PSSM profiles and SAAPs for predicting FAD binding sites in electron transport proteins. The details of the proposed approach are described as follows.
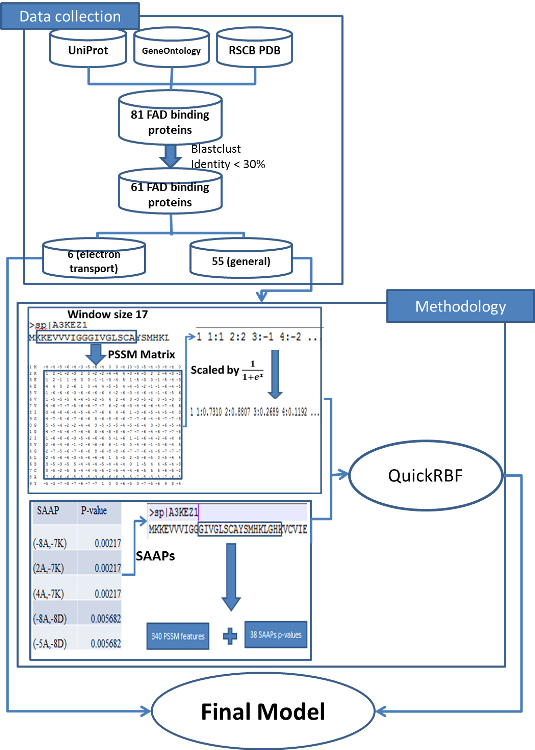
We employed the QuickRBF package to construct RBFN classifiers. The architecture of the RBF network is shown in the below figure. Furthermore, we assigned a constant bandwidth of 5 for each kernel function in the network. We also used all training data as centers. Subsequently, the RBFN classifier was used to identify FAD binding sites according to the output function value.